A real-world case (Biomimetics: space technology)
Here we use a some data produced by precisely modelling a nanostructured solar array inspired by the moth eye. We analyze the relations (found by dcgpy symbolic_regression using mes) between the cell efficiency and 12 different model parameters (thickness, etc..).
The data were generated in the context of the European Space Agency Ariadna study on bio-inspired solar cells (http://www.esa.int/gsp/ACT/projects/diatom/).
The fits were generated evolving 126 populations (size 8) via dcgpy.mes(10000, max_mut=15) on an AMD EPYC 7702 64-Core Processor (~20 minutes to complete)
Data points are pickled into a file named moth_data.pk in the form of a tuple of two arrays Xs, Ys.
The fitted expressions are pickled into a file named result.pk in a list: result[prob_id][0] = fitnesses result[prob_id][1] = chromosomes
This tutorial is informative as it shows the wide diversity and quality of models produced by unconnected evolutions and their difference with respect to the fit error.
Preliminaries
[1]:
# Core imports
import dcgpy # This was compiled in bertha from github HEAD (v1.5 is unreleased)
import pygmo as pg
import numpy as np
# Sympy is nice to have for basic symbolic manipulation.
from sympy import init_printing
from sympy.parsing.sympy_parser import *
init_printing()
# Plotting
%matplotlib inline
import matplotlib.pyplot as plt
# Pickling
import pickle as pk
[2]:
# Load the data to fit
with open("moth_data.pk", "rb") as f:
Xs,Ys = pk.load(f)
# Load the best solutions found running dcgpy.mes(10000, max_mut=15) on 126 separate runs
#with open("eph_3_r_1_c_16_lb_17_sum_diff_mul_diff_exp.pk", "rb") as f:
with open("eph_4_r_1_c_16_lb_15_sum_diff_mul_div_exp_gaussian.pk", "rb") as f:
results = pk.load(f)
kernel_list = ["sum", "diff", "mul", "div", "exp", "gaussian"]
[3]:
# We define the kernel set that was used to produce the runs
ss = dcgpy.kernel_set_double(kernel_list)
[4]:
# Since we have not pickled all the objects used during the experiment, we need to reconstruct the
# udp and the data in a good format.
def reconstruct_udp(prob_id):
X = np.reshape(Xs[prob_id], (len(Xs[prob_id]),1))
Y = np.reshape(Ys[prob_id], (len(Ys[prob_id]),1))
udp = dcgpy.symbolic_regression(points = X, labels = Y, kernels=ss(), n_eph=4, rows =1, cols=16, levels_back=17, multi_objective=False)
return (udp, X, Y)
[5]:
# We also want to know (at a glance) if the experimental setup worked well in terms of MSEs
mses = [min(a[0]) for a in results]
print("Best fit: prob-id ", np.argmin(mses), "| Value: ", mses[np.argmin(mses)][0])
print("Worst fit: prob-id ", np.argmax(mses), "| Value: ", mses[np.argmax(mses)][0])
print("On average: ", sum(mses)/len(mses))
Best fit: prob-id 4 | Value: 3.359546731194255e-06
Worst fit: prob-id 0 | Value: 2.3801558148325692e-05
On average: [1.0948612e-05]
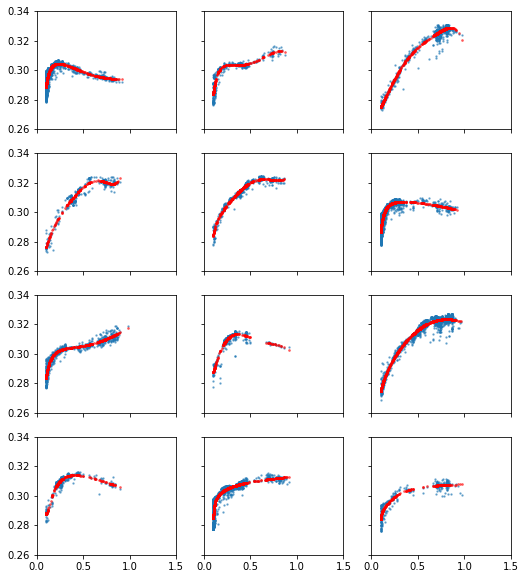
The best fits found
[6]:
# The data and best fits at a glance
fig, axes = plt.subplots(nrows = 4, ncols=3, sharex=True, sharey=True, figsize=(8.5,10))
for i, ax in enumerate(np.reshape(axes,(12))[:12]):
udp, X, Y = reconstruct_udp(i)
ax.scatter(X, Y, s=2.0, alpha=0.5)
best_run = np.argmin(results[i][0])
best_dv = results[i][1][best_run]
Y_pred = udp.predict(X, best_dv)
ax.scatter(X, Y_pred, s=3.0, alpha=0.5, c = 'r')
ax.set_xlim(0, 1.5)
ax.set_ylim(0.26, 0.34)
[6]:
[7]:
# Lets peek into the best fits functional forms. Each of the 12 problems is here
## assigned an id
from IPython.display import display, Math
for prob_id in range(12):
udp, X, Y = reconstruct_udp(prob_id)
best_run= np.argmin(results[prob_id][0])
best_x = results[prob_id][1][best_run]
best_f = results[prob_id][0][best_run]
s = parse_expr(udp.prettier(best_x))[0].simplify()
display("prob_id: {}, err: {}".format(prob_id, best_f[0]), Math(r'{}'.format(s._repr_latex_())))
# NOTE: selecting the fit with the best MSE may not be ideal as best formulas do get unnecessarily complex
'prob_id: 0, err: 2.3801558148325692e-05'
'prob_id: 1, err: 1.5481952594443556e-05'
'prob_id: 2, err: 4.866500757662433e-06'
'prob_id: 3, err: 7.993101895616535e-06'
'prob_id: 4, err: 3.359546731194255e-06'
'prob_id: 5, err: 1.686027444316002e-05'
'prob_id: 6, err: 9.922429392571683e-06'
'prob_id: 7, err: 8.515167695863056e-06'
'prob_id: 8, err: 5.7033656245193685e-06'
'prob_id: 9, err: 4.523905989067585e-06'
'prob_id: 10, err: 1.9017543270061446e-05'
'prob_id: 11, err: 1.1337997121125522e-05'
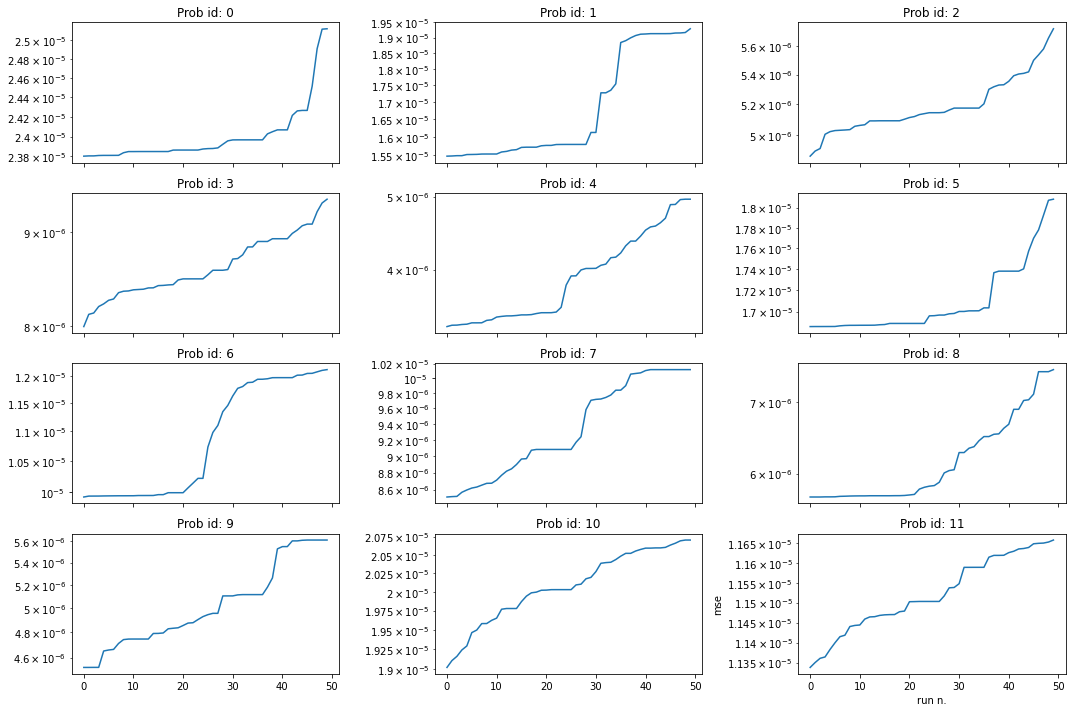
Looking for elegant forms
Some of the best fits found seem to be unnecessarily complex, we here look in the results for more elegant forms with a marginally worse MSA
[8]:
# Lets take for each problem the best 50 fits found and plot the corresponding final MSE
fig, axes = plt.subplots(nrows = 4, ncols=3, sharex=True, figsize=(15,10))
for i, ax in enumerate(np.reshape(axes,(12))[:12]):
a = results[i]
ax.semilogy(sorted(a[0])[:50])
ax.title.set_text("Prob id: " + str(i))
plt.xlabel("run n.")
plt.ylabel("mse")
plt.tight_layout()
# Conclusion: there are many formulas with similar error that can be considered as equally good fits
Showing the best 5 fits
[9]:
for prob_id in range(12):
print("---------------------------------------------------------")
print("Best five fits for id: ", prob_id)
a = results[prob_id]
fits = a[0]
best_idxs = sorted(range(126), key = lambda x: fits[x])
for best_isl in best_idxs[:5]:
best_x = a[1][best_isl]
best_f = a[0][best_isl]
udp, X, Y = reconstruct_udp(prob_id)
s = parse_expr(udp.prettier(best_x))[0].simplify()
display("prob_id: {}, err: {}".format(prob_id, best_f[0]), Math(r'{}'.format(s._repr_latex_())))
print("---------------------------------------------------------")
---------------------------------------------------------
Best five fits for id: 0
'prob_id: 0, err: 2.3801558148325692e-05'
'prob_id: 0, err: 2.380412125841117e-05'
'prob_id: 0, err: 2.3804219347209996e-05'
'prob_id: 0, err: 2.3808190703491017e-05'
'prob_id: 0, err: 2.3809219218852146e-05'
---------------------------------------------------------
---------------------------------------------------------
Best five fits for id: 1
'prob_id: 1, err: 1.5481952594443556e-05'
'prob_id: 1, err: 1.5486489454637892e-05'
'prob_id: 1, err: 1.5494780218604477e-05'
'prob_id: 1, err: 1.549478021860451e-05'
'prob_id: 1, err: 1.5524996126148153e-05'
---------------------------------------------------------
---------------------------------------------------------
Best five fits for id: 2
'prob_id: 2, err: 4.866500757662433e-06'
'prob_id: 2, err: 4.8981961254319396e-06'
'prob_id: 2, err: 4.9140689166325595e-06'
'prob_id: 2, err: 5.004121929538379e-06'
'prob_id: 2, err: 5.0208570048206605e-06'
---------------------------------------------------------
---------------------------------------------------------
Best five fits for id: 3
'prob_id: 3, err: 7.993101895616535e-06'
'prob_id: 3, err: 8.116838729621195e-06'
'prob_id: 3, err: 8.13057337419324e-06'
'prob_id: 3, err: 8.19691765672383e-06'
'prob_id: 3, err: 8.224153596665173e-06'
---------------------------------------------------------
---------------------------------------------------------
Best five fits for id: 4
'prob_id: 4, err: 3.359546731194255e-06'
'prob_id: 4, err: 3.3734252967348888e-06'
'prob_id: 4, err: 3.3746957690291797e-06'
'prob_id: 4, err: 3.3811044286786395e-06'
'prob_id: 4, err: 3.3842683416733736e-06'
---------------------------------------------------------
---------------------------------------------------------
Best five fits for id: 5
'prob_id: 5, err: 1.686027444316002e-05'
'prob_id: 5, err: 1.686067254813111e-05'
'prob_id: 5, err: 1.686069307097475e-05'
'prob_id: 5, err: 1.6860856181522074e-05'
'prob_id: 5, err: 1.686098183474468e-05'
---------------------------------------------------------
---------------------------------------------------------
Best five fits for id: 6
'prob_id: 6, err: 9.922429392571683e-06'
'prob_id: 6, err: 9.936575700754542e-06'
'prob_id: 6, err: 9.936942645025763e-06'
'prob_id: 6, err: 9.937353947520078e-06'
'prob_id: 6, err: 9.938981126078224e-06'
---------------------------------------------------------
---------------------------------------------------------
Best five fits for id: 7
'prob_id: 7, err: 8.515167695863056e-06'
'prob_id: 7, err: 8.519576231706857e-06'
'prob_id: 7, err: 8.522266408037304e-06'
'prob_id: 7, err: 8.569811491073491e-06'
'prob_id: 7, err: 8.596454393800356e-06'
---------------------------------------------------------
---------------------------------------------------------
Best five fits for id: 8
'prob_id: 8, err: 5.7033656245193685e-06'
'prob_id: 8, err: 5.703378409998452e-06'
'prob_id: 8, err: 5.7033784099984615e-06'
'prob_id: 8, err: 5.705047245789752e-06'
'prob_id: 8, err: 5.705078353147797e-06'
---------------------------------------------------------
---------------------------------------------------------
Best five fits for id: 9
'prob_id: 9, err: 4.523905989067585e-06'
'prob_id: 9, err: 4.523956586681867e-06'
'prob_id: 9, err: 4.524279590137839e-06'
'prob_id: 9, err: 4.524279590138164e-06'
'prob_id: 9, err: 4.651129097622205e-06'
---------------------------------------------------------
---------------------------------------------------------
Best five fits for id: 10
'prob_id: 10, err: 1.9017543270061446e-05'
'prob_id: 10, err: 1.9104544589813626e-05'
'prob_id: 10, err: 1.9158737383062652e-05'
'prob_id: 10, err: 1.9240171327253178e-05'
'prob_id: 10, err: 1.9291883990542647e-05'
---------------------------------------------------------
---------------------------------------------------------
Best five fits for id: 11
'prob_id: 11, err: 1.1337997121125522e-05'
'prob_id: 11, err: 1.135016956476325e-05'
'prob_id: 11, err: 1.1360665073366816e-05'
'prob_id: 11, err: 1.1364062900679142e-05'
'prob_id: 11, err: 1.138267448015574e-05'
---------------------------------------------------------