expression (dCGP)
This class represents a Cartesian Genetic Program. Since that is, essentially, an artificial genetic encoding for a mathematical expression, we named the templated class expression. The class template can be instantiated using the types double or gdual<T>. In the case of double, the class would basically reproduce a canonical CGP expression. In the case of gdual<T> the class would operate in the differential algebra of truncated Taylor polynomials with coefficients in T, and thus provide also any order derivative information on the program (i.e. the Taylor expansion of the program output with respect to its inputs).
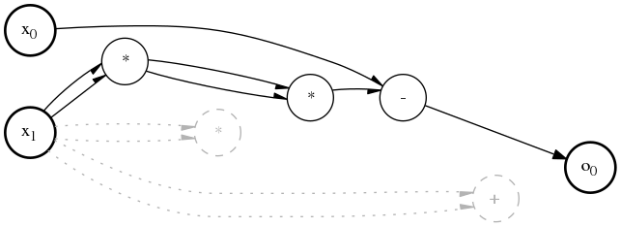
A dCGP expression
-
template<typename T>
class expression A dCGP expression.
This class represents a mathematical expression as encoded using CGP and contains algorithms to compute its value (numerical and symbolical) and its derivatives as well as to mutate the expression.
- Template Parameters
T – expression type. Can be double, or a gdual type.
Subclassed by dcgp::expression_weighted< T >
Public Types
Public Functions
-
inline expression(unsigned n, unsigned m, unsigned r, unsigned c, unsigned l, std::vector<unsigned> arity, std::vector<kernel<T>> f, unsigned n_eph, unsigned seed = dcgp::random_device::next())
Constructor.
Constructs a dCGP expression with variable arity
- Parameters
n – [in] number of inputs (independent variables).
m – [in] number of outputs (dependent variables).
r – [in] number of rows of the cartesian representation of the expression as an acyclic graph.
c – [in] number of columns of the cartesian representation of the expression as an acyclic graph.
l – [in] number of levels-back allowed. This, essentially, controls the minimum number of allowed operations in the formula. If uncertain set it to c + 1
arity – [in] arities of the basis functions for each column.
f – [in] function set. An std::vector of dcgp::kernel<expression::type>.
n_eph – [in] Number of ephemeral constants. Their values and their symbols can be set via the dedicate methods.
seed – [in] seed for the random number generator (initial expression and mutations depend on this).
-
inline expression(unsigned n = 1u, unsigned m = 1u, unsigned r = 1u, unsigned c = 1u, unsigned l = 1u, unsigned arity = 1u, std::vector<kernel<T>> f = kernel_set<T>({"sum"})(), unsigned n_eph = 0u, unsigned seed = dcgp::random_device::next())
Constructor.
Constructs a dCGP expression with uniform arity
- Parameters
n – [in] number of inputs (independent variables).
m – [in] number of outputs (dependent variables).
r – [in] number of rows of the cartesian representation of the expression as an acyclic graph.
c – [in] number of columns of the cartesian representation of the expression as an acyclic graph.
l – [in] number of levels-back allowed. This, essentially, controls the minimum number of allowed operations in the formula. If uncertain set it to c + 1
arity – [in] arity of the basis functions.
f – [in] function set. An std::vector of dcgp::kernel<expression::type>.
n_eph – [in] Number of ephemeral constants. Their values and their symbols can be set via the dedicate methods.
seed – [in] seed for the random number generator (initial expression and mutations depend on this).
-
inline virtual ~expression()
Virtual destructor.
-
expression(const expression&) = default
Copy constructor.
-
expression(expression&&) = default
Copy assignment operator.
-
expression &operator=(const expression&) = default
Move constructor.
-
expression &operator=(expression&&) = default
Move assignment operator.
-
inline virtual std::vector<T> operator()(const std::vector<T> &point) const
Evaluates the dCGP expression.
This evaluates the dCGP expression.
- Parameters
[point] – an std::vector containing the values where the dCGP expression has to be computed (doubles, gduals or strings)
- Returns
The value of the function (an std::vector)
-
inline std::vector<double> operator()(const std::initializer_list<double> &in) const
Evaluates the dCGP expression (from initializer list)
This evaluates the dCGP expression from an initializer list.
- Parameters
in – [in] an initializer list containing the values where the dCGP expression has to be computed (doubles, gduals or strings)
- Returns
The value of the function (an std::vector)
-
inline virtual std::vector<std::string> operator()(const std::vector<std::string> &in) const
Evaluates the dCGP expression (symbolic)
This evaluates the symbolic form of a dCGP expression from symbols.
- Parameters
in – [in] an initializer list containing the symbols to use to construct the dCGP expression.
- Returns
The value of the function (an std::vector)
-
inline std::vector<std::string> operator()(const std::initializer_list<std::string> &in) const
Evaluates the dCGP expression (symbolic from initializer list)
This evaluates the symbolic form of a dCGP expression from an initializer list of symbols.
- Parameters
in – [in] an initializer list containing the symbols to use to construct the dCGP expression.
- Returns
The value of the function (an std::vector)
-
inline T loss(const std::vector<T> &point, const std::vector<T> &prediction, loss_type loss_e) const
Evaluates the model loss (single data point)
Returns the model loss over a single point of data of the dCGP output.
- Parameters
[point] – The input data (single point)
[prediction] – The predicted output (single point)
[loss_e] – The loss type. Can be “MSE” for Mean Square Error (regression) or “CE” for Cross Entropy (classification)
- Returns
the computed loss
-
inline T loss(const std::vector<std::vector<T>> &points, const std::vector<std::vector<T>> &labels, const std::string &loss_s, unsigned parallel = 0u) const
Evaluates the model loss (on a batch)
Evaluates the model loss over a batch.
- Parameters
[points] – The input data (a batch).
[labels] – The predicted outputs (a batch).
[loss_s] – The loss type. Can be “MSE” for Mean Square Error (regression) or “CE” for Cross Entropy (classification)
[parallel] – sets the grain for parallelism. 0 -> no parallelism n -> divides the data into n parts and evaluates them in parallel threads.
- Returns
the loss
-
inline void set(const std::vector<unsigned> &xu)
Sets the chromosome.
Sets a given chromosome as genotype for the expression and updates the active nodes and active genes information accordingly
- Parameters
xu – [in] the new cromosome
- Throws
std::invalid_argument – if the chromosome is out of bounds or has the wrong size.
-
template<class InputIt>
inline void set_from_range(InputIt begin, InputIt end) Sets the chromosome from range.
Sets a given chromosome as genotype for the expression and updates the active nodes and active genes information accordingly
- Parameters
begin – [in] iterator to the first element of the range
end – [in] iterator to the end element of the range
- Throws
std::invalid_argument – if the chromosome is out of bounds or has the wrong size.
-
inline void set_f_gene(unsigned node_id, unsigned f_id)
Sets the function gene of a node.
Sets for a valid node (i.e. not an input node) a new kernel
- Parameters
node_id – [in] the id of the node
f_id – [in] the id of the kernel
- Throws
std::invalid_argument – if the node_id or f_id are invalid.
-
inline void set_eph_val(const std::vector<T> &eph_val)
Sets the values of ephemeral constants.
Sets the values of ephemeral constants
- Parameters
eph_val – [in] the values of the ephemeral constants.
- Throws
std::invalid_argument – if the size of eph_val is not equal to the number of ephemeral constants.
-
inline void set_eph_symb(const std::vector<std::string> &eph_symb)
Sets the values of ephemeral constants.
Sets the values of ephemeral constants
- Parameters
eph_symb – [in] the symbols to use for the ephemeral constants.
- Throws
std::invalid_argument – if the size of eph_symb is not equal to the number of ephemeral constants.
-
inline const std::vector<T> &get_eph_val() const
Gets the values of ephemeral constants.
Gets the values of ephemeral constants
- Returns
the values of ephemeral constants.
-
inline const std::vector<std::string> &get_eph_symb() const
Gets the symbols of ephemeral constants.
Gets the symbols of ephemeral constants
- Returns
the symbols of ephemeral constants.
-
inline const std::vector<unsigned> &get() const
Gets the chromosome.
Gets the chromosome encoding the current expression
- Returns
The chromosome
-
inline const std::vector<unsigned> &get_lb() const
Gets the lower bounds.
Gets the lower bounds for the genes
- Returns
An std::vector containing the lower bound for each gene
-
inline const std::vector<unsigned> &get_ub() const
Gets the upper bounds.
Gets the upper bounds for the genes
- Returns
An std::vector containing the upper bound for each gene
-
inline const std::vector<unsigned> &get_active_genes() const
Gets the active genes.
Gets the idx of the active genes in the current chromosome (numbering is from 0)
- Returns
An std::vector containing the idx of the active genes in the current chromosome
-
inline const std::vector<unsigned> &get_active_nodes() const
Gets the active nodes.
Gets the idx of the active nodes in the current chromosome. The numbering starts from 0 at the first input node to then follow PPSN tutorial from Miller
- Returns
An std::vector containing the idx of the active nodes
-
inline unsigned get_n() const
Gets the number of inputs.
Gets the number of inputs of the dCGP expression
- Returns
the number of inputs
-
inline unsigned get_m() const
Gets the number of outputs.
Gets the number of outputs of the dCGP expression
- Returns
the number of outputs
-
inline unsigned get_r() const
Gets the number of rows.
Gets the number of rows of the dCGP expression
- Returns
the number of rows
-
inline unsigned get_c() const
Gets the number of columns.
Gets the number of columns of the dCGP expression
- Returns
the number of columns
-
inline unsigned get_l() const
Gets the number of levels-back.
Gets the number of levels-back allowed for the dCGP expression
- Returns
the number of levels-back
-
inline const std::vector<unsigned> &get_arity() const
Gets the arity.
Gets the arity of the basis functions of the dCGP expression
- Returns
the arity
-
inline unsigned get_arity(unsigned node_id) const
Gets the arity of a particular node.
Gets the arity of a particular node
- Parameters
node_id – [in] id of the node
- Returns
the arity of that node
-
inline const std::vector<kernel<T>> &get_f() const
Gets the function set.
Gets the set of functions used in the dCGP expression
- Returns
an std::vector of kernels
-
inline const std::vector<unsigned> &get_gene_idx() const
Gets gene_idx.
Gets gene_idx, a vector containing the indexes in the chromosome where nodes start expressing.
- Returns
an std::vector containing the indexes of the chromosome expressing each node
-
inline void mutate(unsigned idx)
Mutates randomly one gene.
Mutates exactly one gene within its allowed bounds.
- Parameters
idx – [in] index of the gene to me mutated
- Throws
std::invalid_argument – if
idxis too large
-
inline void mutate(std::vector<unsigned> idxs)
Mutates multiple genes at once.
Mutates multiple genes within their allowed bounds.
- Parameters
idxs – [in] vector of indexes of the genes to me mutated
- Throws
std::invalid_argument – if
idxis too large
-
inline void mutate_random(unsigned N)
Mutates N random genes.
Mutates a specified number of random genes within their bounds
- Parameters
N – [in] number of genes to be mutated
-
inline void mutate_inactive(unsigned N = 1u)
Mutates inactive genes randomly up to
N.Mutates inactive random genes within their bounds up to
N. The guarantee to actually mutate N would cost and is deemed unnecessary.- Parameters
N – [in] maximum number of inactive genes to be mutated
-
inline void mutate_active(unsigned N = 1u)
Mutates active genes.
Mutates
Nactive genes within their allowed bounds. The mutation can affect function genes, input genes and output genes.- Parameters
N – [in] Number of active genes to be mutated
-
inline void mutate_active_fgene(unsigned N = 1u)
Mutates active function genes.
Mutates
Nactive function genes within their allowed bounds.- Parameters
N – [in] Number of active function genes to be mutated
-
inline void mutate_active_cgene(unsigned N = 1u)
Mutates active connection genes.
Mutates
Nactive connection genes within their allowed bounds.- Parameters
N – [in] Number of active connection genes to be mutated
-
inline void mutate_ogene(unsigned N = 1u)
Mutates active output genes.
Mutates
Ntimes random active output genes within their allowed bounds.- Parameters
N – [in] Number of output genes to be mutated
-
inline void seed(long seed)
Sets the internal seed.
Sets the internal seed used to perform mutations and other things.
-
inline bool is_active_node(const unsigned node_id) const
Checks if a given node is active.
- Parameters
node_id – [in] the node to be checked
- Returns
True if the node node_id is active in the CGP expression.
-
inline bool is_active_gene(const unsigned idx) const
Checks if a given gene is active.
- Parameters
idx – [in] the idx of the gene to be checked
- Returns
True if the gene idx is active in the CGP expression.
-
inline void set_phenotype_correction(pc_fun_type pc)
Sets the phenotype correction.
A phenotype correction is a correction applied to the expression output that depends on the expression itself and on its inputs. Indicating with g the expression, the overall output, after a phenotype expression is applied, will be of the generic form y = pc(x, g)
- Parameters
pc – callable to be applied to the CGP expression.
-
inline void unset_phenotype_correction()
Unsets the phenotype correction.
Friends
-
inline friend std::ostream &operator<<(std::ostream &os, const expression &d)
Overloaded stream operator.
Will return a formatted string containing a human readable representation of the class
- Returns
std::string containing a human-readable representation of the problem.