expression (dCGP)
Three types of expression are currently available in python. They differ from the type they operate upon: double, gdual, vectorized gdual.
expression_double
- class dcgpy.expression_double(inputs, outputs, rows, cols, levels_back, arity = 2, kernels, n_eph = 0, seed = randint)
A CGP expression
Constructs a CGP expression operating on double
- Parameters
inputs (
int) – number of inputsoutputs (
int) – number of outputsrows (
int) – number of rows of the cartesian representation of the expression as an acyclic graph.cols (
int) – number of columns of the cartesian representation of the expression as an acyclic graph.levels_back (
int) – number of levels-back allowed. This, essentially, controls the minimum number of allowed operations in the formula. If uncertain set it to cols + 1arity (
intonlist) – arity of the kernels. Assumed equal for all columns unless its specified by a list. The list must contain a number of entries equal to the number of columns.kernels (
List[dcgpy.kernel_double]) – kernel functionsn_eph (int) – Number of ephemeral constants. Their values and their symbols can be set via dedicated methods.
seed (
int) – random seed to generate mutations and chromosomes
- eph_val
Values of the ephemeral constants.
- Type
list(
`)double`
- eph_symb
Symbols used for the ephemeral constants.
- Type
list(
str)
Examples:
>>> from dcgpy import * >>> dcgp = expression_double(1,1,1,10,11,2,kernel_set_double(["sum","diff","mul","div"])(), 0, 32) >>> print(dcgp) ... >>> num_out = dcgp([in]) >>> sym_out = dcgp(["x"])
- get()
Gets the expression chromosome
- get_active_genes()
Gets the idx of the active genes in the current chromosome (numbering is from 0)
- get_active_nodes()
Gets the idx of the active nodes in the current chromosome
- get_arity()
- get_arity(node_id) None
Gets the arity of the basis functions of the dCGP expression. Either the whole vector or that of a single node.
- get_cols()
Gets the number of columns of the dCGP expression
- get_f()
Gets the kernel functions
- get_gene_idx()
Gets a vector containing the indexes in the chromosome where each node starts to be expressed.
- get_lb()
Gets the lower bounds of the chromosome
- get_levels_back()
Gets the number of levels-back allowed for the dCGP expression
- get_m()
Gets the number of outputs of the dCGP expression
- get_n()
Gets the number of inputs of the dCGP expression
- get_rows()
Gets the number of rows of the dCGP expression
- get_ub()
Gets the upper bounds of the chromosome
- loss(points, labels, loss_type)
Computes the loss of the model on the data
- Parameters
points (2D NumPy float array or
list of listsoffloat) – the input datalabels (2D NumPy float array or
list of listsoffloat) – the output labels (supervised signal)loss_type (
str) – the loss, one of “MSE” for Mean Square Error and “CE” for Cross-Entropy.
- Raises
ValueError – if points or labels are malformed or if loss_type is not one of the available types.
- mutate(idxs)
Mutates multiple genes within their allowed bounds.
- Parameters
idxs (
int) – indexes of the genes to me mutatedidxs – indexes of the single gene to me mutated
- Raises
ValueError – if the index of a gene is out of bounds
mutate(idxs)
Mutates multiple genes within their allowed bounds.
- Parameters
idxs (
int) – indexes of the genes to me mutatedidxs – indexes of the single gene to me mutated
- Raises
ValueError – if the index of a gene is out of bounds
- mutate_active(N=1)
Mutates N randomly selected active genes within their allowed bounds
- mutate_active_cgene(N=1)
Mutates N randomly selected active connections within their allowed bounds
- mutate_active_fgene(N=1)
Mutates N randomly selected active function genes within their allowed bounds
- mutate_ogene(N=1)
Mutates N randomly selected output genes connection within their allowed bounds
- mutate_random(N=1)
Mutates N randomly selected genes within its allowed bounds
- set(chromosome)
Sets the chromosome.
- Parameters
chromosome (
List[int]) – the new chromosome- Raises
ValueError – if the the chromosome is incompatible with the expression (n.inputs, n.outputs, levels-back, etc.)
- set_f_gene(node_id, f_id)
Sets for a valid node (i.e. not an input node) a new kernel.
- Parameters
node_id (
List[int]) – the node idf_id (
List[int]) – the kernel id
- Raises
ValueError – if the node_id or f_id are incompatible with the expression.
- set_phenotype_correction(pc)
A phenotype correction is a correction applied to the expression output that depends on the expression itself and on its inputs.
Indicating with g the expression, the overall output, after a phenotype expression is applied, will be of the generic for y = pc(x, g)
- Parameters
pc (
callable) – callable to be applied to the CGP expression input x and the encoded expression g. No checks are done on the actual dimensions of the input and or outputs.
Examples:
>>> import dcgpy >>> ex = expression_double(1,1,1,10,11,2,kernel_set_double(["sum","diff","mul","div"])(), 0, 33) >>> def pc(x,g): ... return [g(x)[0]*x[0]] >>> ex.set_phenotype_correction(pc)
- simplify(in_sym, subs_weights=False, erc=[])
Simplifies the symbolic output of dCGP expressions
Returns the simplified dCGP expression for each output
Note
This method requires
sympyandpyaudimodules installed in your Python system- Parameters
in_sym (a
List[str]) – input symbols (its length must match the number of inputs)subs_weights (a
bool) – indicates whether to substitute the weights symbols with their valueserc (a
List[float]) – values of the ephemeral random constants (if empty their symbolic representation is used instead)
- Returns
A
List[str]containing the simplified expressions- Raises
ValueError – if the length of in_sym does not match the number of inputs
ValueError – if the length of erc is larger than the number of inputs
ImportError – if modules sympy or pyaudi are not installed in your Python system
Examples
>>> ex = dcgpy.expression_weighted_gdual_double(3,2,3,3,2,2,dcgpy.kernel_set_gdual_double(["sum","diff"])(),0) >>> print(ex.simplify(['x','c0','c1'],True,[1,2])[0]) x + 6
- unset_phenotype_correction()
Unsets the phenotype correction.
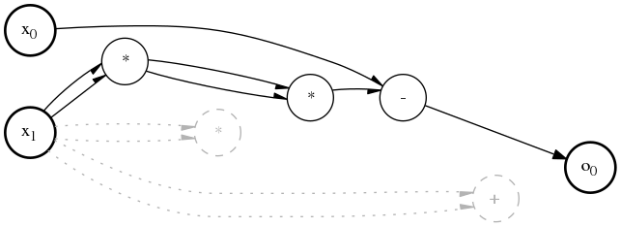
- visualize(in_sym=[], draw_inactive=True, draw_weights=False)
Visualizes the d-CGP expression Visualizes the graph of the d-CGP expression .. note:: This method requires the graphviz` module installed in your Python system
- Parameters
in_sym (a
List[str]) – input symbols. Its length must either match the number of inputs or be zero (to visualize them as x_i)draw_inactive (a
bool) – indicates whether to draw inactive nodesdraw_weights (a
bool) – indicates whether to draw connection weights symbols
- Returns
The
graphviz.Digraphfor the given expression- Raises
ImportError – if module pygraphviz is not installed in your Python system
ValueError – if in_sym is nonempty but its length does not match the number of inputs
Examples
>>> ex = dcgpy.expression_double(2,1,3,3,2,2,dcgpy.kernel_set_double(["sum","diff"])(),0) >>> graph = ex.visualize(['x', 'c'], True, False)
expression_gdual_double
- class dcgpy.expression_gdual_double(inputs, outputs, rows, cols, levels_back, arity = 2, kernels, n_eph = 0, seed = randint)
A CGP expression
Constructs a CGP expression operating on gdual_double
- Parameters
inputs (
int) – number of inputsoutputs (
int) – number of outputsrows (
int) – number of rows of the cartesian representation of the expression as an acyclic graph.cols (
int) – number of columns of the cartesian representation of the expression as an acyclic graph.levels_back (
int) – number of levels-back allowed. This, essentially, controls the minimum number of allowed operations in the formula. If uncertain set it to cols + 1arity (
intonlist) – arity of the kernels. Assumed equal for all columns unless its specified by a list. The list must contain a number of entries equal to the number of columns.kernels (
List[dcgpy.kernel_gdual_double]) – kernel functionsn_eph (int) – Number of ephemeral constants. Their values and their symbols can be set via dedicated methods.
seed (
int) – random seed to generate mutations and chromosomes
- eph_val
Values of the ephemeral constants.
- Type
list(
`)gdual_double`
- eph_symb
Symbols used for the ephemeral constants.
- Type
list(
str)
Examples:
>>> from dcgpy import * >>> dcgp = expression_gdual_double(1,1,1,10,11,2,kernel_set_gdual_double(["sum","diff","mul","div"])(), 0, 32) >>> print(dcgp) ... >>> num_out = dcgp([in]) >>> sym_out = dcgp(["x"])
- get()
Gets the expression chromosome
- get_active_genes()
Gets the idx of the active genes in the current chromosome (numbering is from 0)
- get_active_nodes()
Gets the idx of the active nodes in the current chromosome
- get_arity()
- get_arity(node_id) None
Gets the arity of the basis functions of the dCGP expression. Either the whole vector or that of a single node.
- get_cols()
Gets the number of columns of the dCGP expression
- get_f()
Gets the kernel functions
- get_gene_idx()
Gets a vector containing the indexes in the chromosome where each node starts to be expressed.
- get_lb()
Gets the lower bounds of the chromosome
- get_levels_back()
Gets the number of levels-back allowed for the dCGP expression
- get_m()
Gets the number of outputs of the dCGP expression
- get_n()
Gets the number of inputs of the dCGP expression
- get_rows()
Gets the number of rows of the dCGP expression
- get_ub()
Gets the upper bounds of the chromosome
- loss(points, labels, loss_type)
Computes the loss of the model on the data
- Parameters
points (2D NumPy float array or
list of listsoffloat) – the input datalabels (2D NumPy float array or
list of listsoffloat) – the output labels (supervised signal)loss_type (
str) – the loss, one of “MSE” for Mean Square Error and “CE” for Cross-Entropy.
- Raises
ValueError – if points or labels are malformed or if loss_type is not one of the available types.
- mutate(idxs)
Mutates multiple genes within their allowed bounds.
- Parameters
idxs (
int) – indexes of the genes to me mutatedidxs – indexes of the single gene to me mutated
- Raises
ValueError – if the index of a gene is out of bounds
mutate(idxs)
Mutates multiple genes within their allowed bounds.
- Parameters
idxs (
int) – indexes of the genes to me mutatedidxs – indexes of the single gene to me mutated
- Raises
ValueError – if the index of a gene is out of bounds
- mutate_active(N=1)
Mutates N randomly selected active genes within their allowed bounds
- mutate_active_cgene(N=1)
Mutates N randomly selected active connections within their allowed bounds
- mutate_active_fgene(N=1)
Mutates N randomly selected active function genes within their allowed bounds
- mutate_ogene(N=1)
Mutates N randomly selected output genes connection within their allowed bounds
- mutate_random(N=1)
Mutates N randomly selected genes within its allowed bounds
- set(chromosome)
Sets the chromosome.
- Parameters
chromosome (
List[int]) – the new chromosome- Raises
ValueError – if the the chromosome is incompatible with the expression (n.inputs, n.outputs, levels-back, etc.)
- set_f_gene(node_id, f_id)
Sets for a valid node (i.e. not an input node) a new kernel.
- Parameters
node_id (
List[int]) – the node idf_id (
List[int]) – the kernel id
- Raises
ValueError – if the node_id or f_id are incompatible with the expression.
- set_phenotype_correction(pc)
A phenotype correction is a correction applied to the expression output that depends on the expression itself and on its inputs.
Indicating with g the expression, the overall output, after a phenotype expression is applied, will be of the generic for y = pc(x, g)
- Parameters
pc (
callable) – callable to be applied to the CGP expression input x and the encoded expression g. No checks are done on the actual dimensions of the input and or outputs.
Examples:
>>> import dcgpy >>> ex = expression_double(1,1,1,10,11,2,kernel_set_double(["sum","diff","mul","div"])(), 0, 33) >>> def pc(x,g): ... return [g(x)[0]*x[0]] >>> ex.set_phenotype_correction(pc)
- simplify(in_sym, subs_weights=False, erc=[])
Simplifies the symbolic output of dCGP expressions
Returns the simplified dCGP expression for each output
Note
This method requires
sympyandpyaudimodules installed in your Python system- Parameters
in_sym (a
List[str]) – input symbols (its length must match the number of inputs)subs_weights (a
bool) – indicates whether to substitute the weights symbols with their valueserc (a
List[float]) – values of the ephemeral random constants (if empty their symbolic representation is used instead)
- Returns
A
List[str]containing the simplified expressions- Raises
ValueError – if the length of in_sym does not match the number of inputs
ValueError – if the length of erc is larger than the number of inputs
ImportError – if modules sympy or pyaudi are not installed in your Python system
Examples
>>> ex = dcgpy.expression_weighted_gdual_double(3,2,3,3,2,2,dcgpy.kernel_set_gdual_double(["sum","diff"])(),0) >>> print(ex.simplify(['x','c0','c1'],True,[1,2])[0]) x + 6
- unset_phenotype_correction()
Unsets the phenotype correction.
- visualize(in_sym=[], draw_inactive=True, draw_weights=False)
Visualizes the d-CGP expression Visualizes the graph of the d-CGP expression .. note:: This method requires the graphviz` module installed in your Python system
- Parameters
in_sym (a
List[str]) – input symbols. Its length must either match the number of inputs or be zero (to visualize them as x_i)draw_inactive (a
bool) – indicates whether to draw inactive nodesdraw_weights (a
bool) – indicates whether to draw connection weights symbols
- Returns
The
graphviz.Digraphfor the given expression- Raises
ImportError – if module pygraphviz is not installed in your Python system
ValueError – if in_sym is nonempty but its length does not match the number of inputs
Examples
>>> ex = dcgpy.expression_double(2,1,3,3,2,2,dcgpy.kernel_set_double(["sum","diff"])(),0) >>> graph = ex.visualize(['x', 'c'], True, False)
expression_gdual_vdouble
- class dcgpy.expression_gdual_vdouble(inputs, outputs, rows, cols, levels_back, arity = 2, kernels, n_eph = 0, seed = randint)
A CGP expression
Constructs a CGP expression operating on gdual_vdouble
- Parameters
inputs (
int) – number of inputsoutputs (
int) – number of outputsrows (
int) – number of rows of the cartesian representation of the expression as an acyclic graph.cols (
int) – number of columns of the cartesian representation of the expression as an acyclic graph.levels_back (
int) – number of levels-back allowed. This, essentially, controls the minimum number of allowed operations in the formula. If uncertain set it to cols + 1arity (
intonlist) – arity of the kernels. Assumed equal for all columns unless its specified by a list. The list must contain a number of entries equal to the number of columns.kernels (
List[dcgpy.kernel_gdual_vdouble]) – kernel functionsn_eph (int) – Number of ephemeral constants. Their values and their symbols can be set via dedicated methods.
seed (
int) – random seed to generate mutations and chromosomes
- eph_val
Values of the ephemeral constants.
- Type
list(
`)gdual_vdouble`
- eph_symb
Symbols used for the ephemeral constants.
- Type
list(
str)
Examples:
>>> from dcgpy import * >>> dcgp = expression_gdual_vdouble(1,1,1,10,11,2,kernel_set_gdual_vdouble(["sum","diff","mul","div"])(), 0, 32) >>> print(dcgp) ... >>> num_out = dcgp([in]) >>> sym_out = dcgp(["x"])
- get()
Gets the expression chromosome
- get_active_genes()
Gets the idx of the active genes in the current chromosome (numbering is from 0)
- get_active_nodes()
Gets the idx of the active nodes in the current chromosome
- get_arity()
- get_arity(node_id) None
Gets the arity of the basis functions of the dCGP expression. Either the whole vector or that of a single node.
- get_cols()
Gets the number of columns of the dCGP expression
- get_f()
Gets the kernel functions
- get_gene_idx()
Gets a vector containing the indexes in the chromosome where each node starts to be expressed.
- get_lb()
Gets the lower bounds of the chromosome
- get_levels_back()
Gets the number of levels-back allowed for the dCGP expression
- get_m()
Gets the number of outputs of the dCGP expression
- get_n()
Gets the number of inputs of the dCGP expression
- get_rows()
Gets the number of rows of the dCGP expression
- get_ub()
Gets the upper bounds of the chromosome
- loss(points, labels, loss_type)
Computes the loss of the model on the data
- Parameters
points (2D NumPy float array or
list of listsoffloat) – the input datalabels (2D NumPy float array or
list of listsoffloat) – the output labels (supervised signal)loss_type (
str) – the loss, one of “MSE” for Mean Square Error and “CE” for Cross-Entropy.
- Raises
ValueError – if points or labels are malformed or if loss_type is not one of the available types.
- mutate(idxs)
Mutates multiple genes within their allowed bounds.
- Parameters
idxs (
int) – indexes of the genes to me mutatedidxs – indexes of the single gene to me mutated
- Raises
ValueError – if the index of a gene is out of bounds
mutate(idxs)
Mutates multiple genes within their allowed bounds.
- Parameters
idxs (
int) – indexes of the genes to me mutatedidxs – indexes of the single gene to me mutated
- Raises
ValueError – if the index of a gene is out of bounds
- mutate_active(N=1)
Mutates N randomly selected active genes within their allowed bounds
- mutate_active_cgene(N=1)
Mutates N randomly selected active connections within their allowed bounds
- mutate_active_fgene(N=1)
Mutates N randomly selected active function genes within their allowed bounds
- mutate_ogene(N=1)
Mutates N randomly selected output genes connection within their allowed bounds
- mutate_random(N=1)
Mutates N randomly selected genes within its allowed bounds
- set(chromosome)
Sets the chromosome.
- Parameters
chromosome (
List[int]) – the new chromosome- Raises
ValueError – if the the chromosome is incompatible with the expression (n.inputs, n.outputs, levels-back, etc.)
- set_f_gene(node_id, f_id)
Sets for a valid node (i.e. not an input node) a new kernel.
- Parameters
node_id (
List[int]) – the node idf_id (
List[int]) – the kernel id
- Raises
ValueError – if the node_id or f_id are incompatible with the expression.
- set_phenotype_correction(pc)
A phenotype correction is a correction applied to the expression output that depends on the expression itself and on its inputs.
Indicating with g the expression, the overall output, after a phenotype expression is applied, will be of the generic for y = pc(x, g)
- Parameters
pc (
callable) – callable to be applied to the CGP expression input x and the encoded expression g. No checks are done on the actual dimensions of the input and or outputs.
Examples:
>>> import dcgpy >>> ex = expression_double(1,1,1,10,11,2,kernel_set_double(["sum","diff","mul","div"])(), 0, 33) >>> def pc(x,g): ... return [g(x)[0]*x[0]] >>> ex.set_phenotype_correction(pc)
- simplify(in_sym, subs_weights=False, erc=[])
Simplifies the symbolic output of dCGP expressions
Returns the simplified dCGP expression for each output
Note
This method requires
sympyandpyaudimodules installed in your Python system- Parameters
in_sym (a
List[str]) – input symbols (its length must match the number of inputs)subs_weights (a
bool) – indicates whether to substitute the weights symbols with their valueserc (a
List[float]) – values of the ephemeral random constants (if empty their symbolic representation is used instead)
- Returns
A
List[str]containing the simplified expressions- Raises
ValueError – if the length of in_sym does not match the number of inputs
ValueError – if the length of erc is larger than the number of inputs
ImportError – if modules sympy or pyaudi are not installed in your Python system
Examples
>>> ex = dcgpy.expression_weighted_gdual_double(3,2,3,3,2,2,dcgpy.kernel_set_gdual_double(["sum","diff"])(),0) >>> print(ex.simplify(['x','c0','c1'],True,[1,2])[0]) x + 6
- unset_phenotype_correction()
Unsets the phenotype correction.
- visualize(in_sym=[], draw_inactive=True, draw_weights=False)
Visualizes the d-CGP expression Visualizes the graph of the d-CGP expression .. note:: This method requires the graphviz` module installed in your Python system
- Parameters
in_sym (a
List[str]) – input symbols. Its length must either match the number of inputs or be zero (to visualize them as x_i)draw_inactive (a
bool) – indicates whether to draw inactive nodesdraw_weights (a
bool) – indicates whether to draw connection weights symbols
- Returns
The
graphviz.Digraphfor the given expression- Raises
ImportError – if module pygraphviz is not installed in your Python system
ValueError – if in_sym is nonempty but its length does not match the number of inputs
Examples
>>> ex = dcgpy.expression_double(2,1,3,3,2,2,dcgpy.kernel_set_double(["sum","diff"])(),0) >>> graph = ex.visualize(['x', 'c'], True, False)